34 Survival analysis
In health sciences we often have data which represent the time until some event occurs. In a clinical trial setting, the primary outcome may be the time from onset of therapy until a well-defined critical event such as death, cancer recurrence, or first occurrence of a particular adverse event. One of the challenges specific to survival data is that the survival times will be unknown for some patients (censored data) for a variety of reasons:
- lost to follow-up during the study period
- withdrawn from the study because of some reason
- the event did not occur by the end of the study
Such data could be referred to as right censored because, on a timeline, the true lifetimes of the patients are to the right of their observed censor times.
Statistical techniques for survival data have been developed that are be able to account for censored observations. In survival analysis, it is generally assumed that censoring is non-informative of survival (being censored or not is not related to the probability of the event occurring).
Survival analysis is a class of statistical methods. One approach to estimate the the survival over time is the Kaplan-Meier procedure which is a non-parametric method (i.e. it does not make any assumptions about the underlying distribution of the data).
When we have finished this Chapter, we should be able to:
34.1 Research question
In a randomized controlled trial designed to find out the efficacy of a new therapy for leukemia, the patients were randomly assigned into two groups (new therapy versus standard therapy). The researchers want to (i) estimate the survival time of patients receiving the new therapy, and (ii) compare the survival curves of the patients receiving the new therapy and patients receiving the standard therapy.
34.2 Packages we need
We need to load the following packages:
34.3 Preparing the data
In a clinical trial, the patients are monitored for the occurrence of a particular event or outcome. In preparing Kaplan-Meier survival analysis that compares different treatments, three variables should be recorded for each patient: (a) the time that is the duration between the beginning of the treatment and the end-point (event of interest or censoring), (b) the status at the end of the survival time (event occurrence or censored data), and (c) the study group such as treatment versus control intervention.
We import the data “leukemia” in R:
library(readxl)
dat <- read_excel(here("data", "leukemia.xlsx"))The dataset has 42 observations (rows) and includes three variables (columns).
time: the survival or censoring time in months
status: indicator whether or not the patient died (1 indicates death and 0 indicates censored observation)
intervention: randomly assigned therapy group with two levels, therapy A (new) or therapy B (standard).
We inspect the data and the type of variables:
glimpse(dat)Rows: 42
Columns: 3
$ time <dbl> 6, 6, 6, 6, 7, 9, 10, 10, 11, 13, 16, 17, 19, 20, 22, 23,…
$ status <dbl> 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, …
$ intervention <chr> "therapy A (new)", "therapy A (new)", "therapy A (new)", …First, we obtain a subset of data with patients receiving the new therapy A :
dat.A <- dat |>
filter(intervention == "therapy A (new)")Next, we run the Surv() function that converts the data to a special format which allows to account for censored observations (i.e. a compiled version of the survival time and status). The arguments we need to provide are the variables time and status of the patients. The times (in months) for the patients receiving the new therapy A follow:
Surv(dat.A$time, dat.A$status) [1] 6+ 6 6 6 7 9+ 10+ 10 11+ 13 16 17+ 19+ 20+ 22 23 25+ 32+ 32+
[20] 34+ 35+NOTE: Censoring times are marked with “+” symbol. The above data show that patients in the new therapy were censored on months 6+, 9+, 10+, 11+, 17+, 19+, 20+, 32+, 32+, 34+, 35+.
34.4 The Kaplan–Meier Product Limit Estimator
Kaplan-Meier estimator, also known as product-limit estimator, can be used to measure the cumulative probability of “surviving” for a certain amount of time after starting the therapy.
34.4.1 Basic concepts
Let
where
The Kaplan–Meier estimator for the survival function
Therefore, the cumulative probability of surviving beyond
This implies that:
34.4.2 K-M analysis for the group of patients in new therapy A
The first step in Kaplan-Meier analysis usually involves the construction of a table with the Kaplan–Meier estimates, so we call the survfit2() function that models the survival probability with the formula Surv ∼ 1 (we use 1 as we have not any grouping variable):
# create an object with the K-M estimates for the patients in the new therapy A
km.A <- survfit2(Surv(time, status) ~ 1, data = dat.A)
# obtain the list of variables included in the Km.A object
names(km.A) [1] "n" "time" "n.risk" "n.event" "n.censor"
[6] "surv" "std.err" "cumhaz" "std.chaz" "type"
[11] "logse" "conf.int" "conf.type" "lower" "upper"
[16] "call" ".Environment"As we can see, the function survfit() returns a list of variables including the number of participants at risk (n.risk), censored (n.censor) and having experienced an event (n.event) as well as the cumulative probability of surviving over time (surv) for the new therapy, among others.
Next, we estimate in R the conditional probability of surviving over time using the Equation 34.1:
# compute the probability of "surviving" over time
prob.A <- round(1-(km.A$n.event/km.A$n.risk), 3)
prob.A [1] 0.857 0.941 1.000 0.933 1.000 0.917 0.909 1.000 1.000 1.000 0.857 0.833
[13] 1.000 1.000 1.000 1.000
Now, we are ready to include all the information in one table (Table 34.1) as follows:
# create a dataframe with all the "survival" variables of interest
tb.A <- data.frame(time = km.A$time, n.risk = km.A$n.risk,
n.event = km.A$n.event, n.censor = km.A$n.censor,
prob.A, surv.A = round(km.A$surv, 3))
# create the table
tb.A |>
gt() |>
cols_label(
time = html("Ordered times (months), t"),
n.risk = html("No. at risk"),
n.event = html("No. of deaths"),
n.censor = html("No. censored"),
prob.A = html("Conditional probability of surviving, P(t)"),
surv.A = html("Cumulative probability of surviving, S(t)")) |>
tab_options(column_labels.font.weight = "bold") |>
cols_align(align = "center") |>
gt_highlight_rows(rows = 3, fill = "lightgrey",
bold_target_only = TRUE,
target_col = c(n.risk, n.censor)) |>
tab_style(style = list(cell_text(weight = "bold")),
locations = cells_body(columns = n.risk, rows = n.risk == 15))| Ordered times (months), t | No. at risk | No. of deaths | No. censored | Conditional probability of surviving, P(t) | Cumulative probability of surviving, S(t) |
|---|---|---|---|---|---|
| 6 | 21 | 3 | 1 | 0.857 | 0.857 |
| 7 | 17 | 1 | 0 | 0.941 | 0.807 |
| 9 | 16 | 0 | 1 | 1.000 | 0.807 |
| 10 | 15 | 1 | 1 | 0.933 | 0.753 |
| 11 | 13 | 0 | 1 | 1.000 | 0.753 |
| 13 | 12 | 1 | 0 | 0.917 | 0.690 |
| 16 | 11 | 1 | 0 | 0.909 | 0.627 |
| 17 | 10 | 0 | 1 | 1.000 | 0.627 |
| 19 | 9 | 0 | 1 | 1.000 | 0.627 |
| 20 | 8 | 0 | 1 | 1.000 | 0.627 |
| 22 | 7 | 1 | 0 | 0.857 | 0.538 |
| 23 | 6 | 1 | 0 | 0.833 | 0.448 |
| 25 | 5 | 0 | 1 | 1.000 | 0.448 |
| 32 | 4 | 0 | 2 | 1.000 | 0.448 |
| 34 | 2 | 0 | 1 | 1.000 | 0.448 |
| 35 | 1 | 0 | 1 | 1.000 | 0.448 |
When there are only censored observations at a particular time such as at month 9, the conditional probability
Only events cause the survival curve to drop. Censoring causes a larger drop for the next event because it reduces the number of patients at risk when that next event occurs.
34.4.3 The Kaplan–Meier curve
The cumulative survival probability is calculated at each time
tb.km.A <- data.frame(tb.A, SE = round(km.A$std.err, 3), LCL = round(km.A$lower, 3), UCL = round(km.A$upper, 3))
tb.km.A |>
select(-n.censor) |>
filter(prob.A != 1) |>
mutate(j = row_number()) |>
relocate(j) |>
gt() |>
cols_label(
time = html("Survival times (months), t<sub>j</sub>"),
n.risk = html("No. at risk, r<sub>j</sup>"),
n.event = html("No. of deaths, d<sub>j</sup>"),
prob.A = html("Conditional probability of surviving, P(t<sub>j</sub>)"),
surv.A = html("Cumulative probability of surviving, S(t<sub>j</sub>)"),
SE = html("Standard Error of S(t<sub>j</sub>)"),
LCL = html("lower 95% CI"),
UCL = html("upper 95% CI")) |>
tab_options(column_labels.font.weight = "bold") |>
cols_align(align = "center") | j | Survival times (months), tj | No. at risk, r j | No. of deaths, d j | Conditional probability of surviving, P(tj) | Cumulative probability of surviving, S(tj) | Standard Error of S(tj) | lower 95% CI | upper 95% CI |
|---|---|---|---|---|---|---|---|---|
| 1 | 6 | 21 | 3 | 0.857 | 0.857 | 0.089 | 0.720 | 1.000 |
| 2 | 7 | 17 | 1 | 0.941 | 0.807 | 0.108 | 0.653 | 0.996 |
| 3 | 10 | 15 | 1 | 0.933 | 0.753 | 0.128 | 0.586 | 0.968 |
| 4 | 13 | 12 | 1 | 0.917 | 0.690 | 0.155 | 0.510 | 0.935 |
| 5 | 16 | 11 | 1 | 0.909 | 0.627 | 0.182 | 0.439 | 0.896 |
| 6 | 22 | 7 | 1 | 0.857 | 0.538 | 0.238 | 0.337 | 0.858 |
| 7 | 23 | 6 | 1 | 0.833 | 0.448 | 0.300 | 0.249 | 0.807 |
From Table 34.2 we can see that the standard error of the S(t) increases over time as a result of estimating the survival probability at later times with less individuals.
Kaplan–Meier analysis is usually presented as a survival curve in addition to tabular form. The K-M curve of the estimated cumulative probability of surviving with 95% confidence interval (lower 95% CI, upper 95% CI) is depicted in Figure 34.2.
km.A |>
ggsurvfit(linewidth = 1, color = "#077E97" ) +
#theme_prism(palette = "winter_bright", base_size = 12) +
add_confidence_interval(fill = "#077E97") +
add_censor_mark(color = "brown", size = 3.5) +
add_risktable(risktable_stats = c("n.risk", "cum.censor", "cum.event")) +
add_quantile(y_value = 0.5, color = "gray50", linewidth = 0.75) +
scale_x_continuous(expand = c(0.018, 0, 0.02, 0),
limits = c(0, 35), breaks = seq(0, 35, 5)) +
scale_y_continuous(expand = c(0.018, 0, 0.05, 0)) +
scale_colour_prism() +
labs(title = "Kaplan-Meier curve for the new therapy A",
x = "Time in months",
y = "Cumulative Survival Probability") +
theme(panel.grid.major.y = element_line(linewidth = 0.02, color = "grey80"))In Figure 34.2, the time post randomization in months is represented on the x-axis and the cumulative survival probability is plotted on the y-axis. The K-M curve is a stepped line, rather than a smooth curve, since it is horizontal when there is no event and drops only at times when a death occurs. Additionally, the shaded region represents the 95% confidence interval of the
- At time
- The survival curve is defined only up to 35 months, the largest of the observation times.
- Censoring times are marked on the K-M curve as “+” symbols.
At the bottom of the graph in Figure 34.2 there is a table that presents, the number of participants at risk, the cumulative number of censored observations, and the cumulative number of patients having experienced an event at specific time points (0, 5, 10, 15, 20, 25, 30 and 35 months).
We can also estimate the median survival time -the time point at which half of the patients have survived- graphically. In Figure 34.2, from the mid-point of the survival axis (Y = 0.50) move horizontally until the curve is crossed, then drop vertically to the time axis (X-axis) to find the corresponding time. In our example, the median survival is approximately 23 months for the new therapy A.
If the survival curve does not drop to 0.50 or below then the median survival time cannot be computed.
If we want to find in R the median time of surviving for patients in the new therapy A, we have to call the km.A survival object:
km.ACall: survfit(formula = Surv(time, status) ~ 1, data = dat.A)
n events median 0.95LCL 0.95UCL
[1,] 21 9 23 16 NAWe observe that the median time equals to 23 months for the patients receiving the new therapy A.
34.5 Comparing survival curves
34.5.1 Graphical comparison
Survival of two or more groups of patients can be compared graphically. In our example, we are interested in comparing the survival curves between the new therapy and the standard therapy.
- First, in
survfit2()function we need to replace ~1 in the model formula with theinterventionvariable (i.e Surv ~ intervention):
- Then, we use the
ggsurvfit()function to dispaly a Kaplan-Meier curve for each therapy (Figure 34.3):
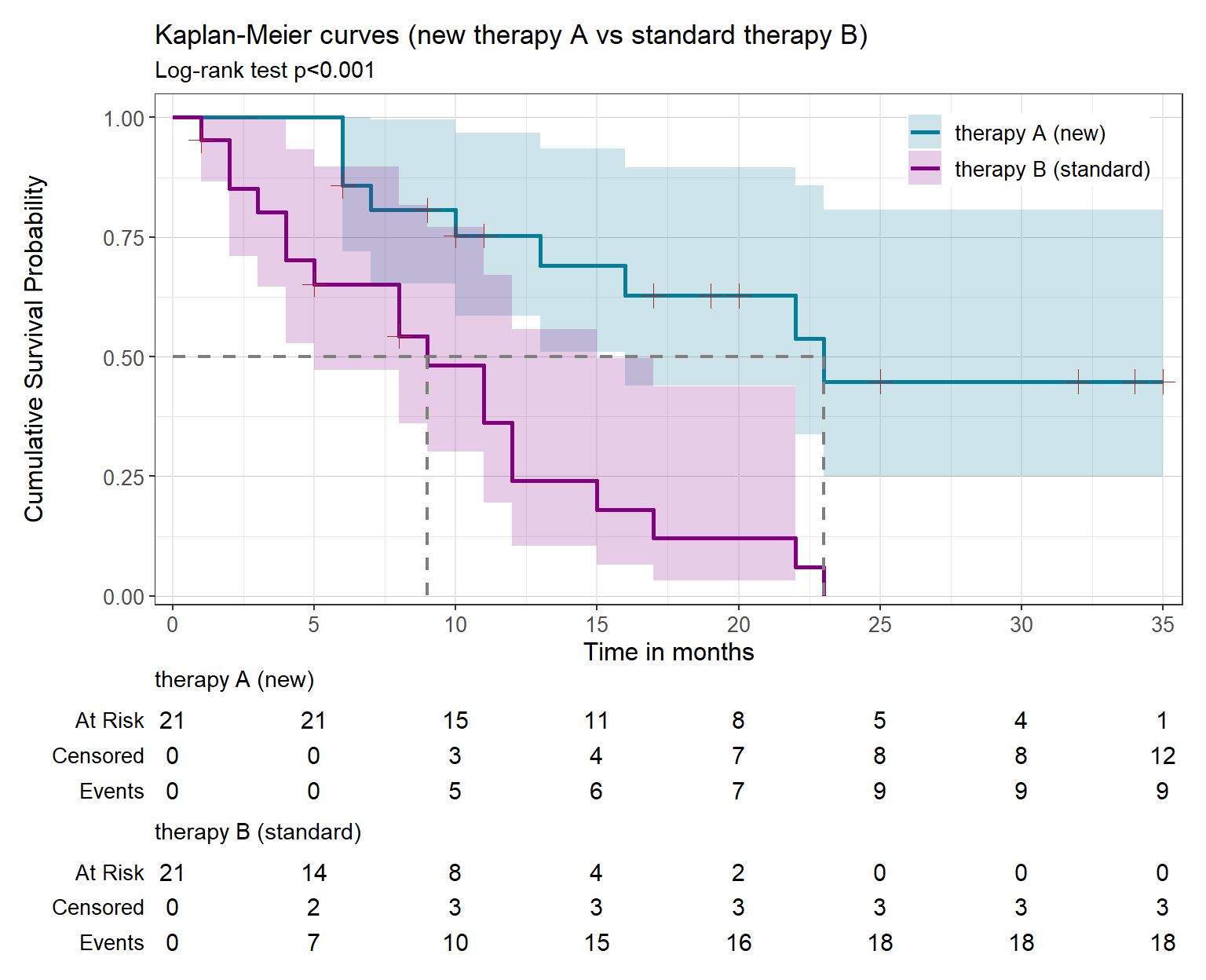
km.AB |>
ggsurvfit(linewidth = 1) +
#theme_prism(palette = "winter_bright", base_size = 12) +
add_confidence_interval() +
add_censor_mark(color = "brown", size = 3.5) +
add_risktable(risktable_stats = c("n.risk", "cum.censor", "cum.event"), size = 4) +
add_quantile(y_value = 0.5, color = "gray50", linewidth = 0.75) +
scale_x_continuous(expand = c(0.018, 0, 0.02, 0), limits = c(0, 35), breaks = seq(0, 35, 5)) +
scale_y_continuous(expand = c(0.018, 0, 0.05, 0)) +
scale_color_manual(values = c("#077E97", "#800080")) +
scale_fill_manual(values = c("#077E97", "#800080")) +
labs(title = "Kaplan-Meier curves (new therapy A vs standard therapy B)",
subtitle = glue::glue("Log-rank test {survfit2_p(km.AB)}"),
x = "Time in months",
y = "Cumulative Survival Probability") +
theme(panel.grid.major.y = element_line(linewidth = 0.02, color = "grey80"),
legend.position = c(0.85, 0.89))K-M plot reveals that patients receiving the new therapy have higher probability of surviving through the whole time period. Particularly, the median survival time for each group can be estimated calling the object km.AB:
# call the km.AB object to find which month corresponds to median survival run
km.ABCall: survfit(formula = Surv(time, status) ~ intervention, data = dat)
n events median 0.95LCL 0.95UCL
intervention=therapy A (new) 21 9 23 16 NA
intervention=therapy B (standard) 21 18 9 5 15The median survival of patients in the new therapy (23 months) is higher than in standard therapy (9 months).
We can plot the cumulative risk function (aka “cumulative incidence” or “cumulative events”), type = "risk". This is the probability of an event, as opposed to the probability of being free from the event (survival).
km.AB |>
ggsurvfit(type = "risk", linewidth = 1) +
#theme_prism(palette = "winter_bright", base_size = 12) +
add_confidence_interval() +
add_censor_mark(color = "brown", size = 3.5) +
add_risktable(risktable_stats = c("n.risk", "cum.censor", "cum.event"), size = 4) +
add_quantile(y_value = 0.5, color = "gray50", linewidth = 0.75) +
scale_x_continuous(expand = c(0.018, 0, 0.02, 0), limits = c(0, 35),
breaks = seq(0, 35, 5)) +
scale_y_continuous(expand = c(0, 0.018, 0.02, 0)) +
scale_color_manual(values = c("#077E97", "#800080")) +
scale_fill_manual(values = c("#077E97", "#800080")) +
labs(title = "Kaplan-Meier curves (new therapy A vs standard therapy B)",
subtitle = glue::glue("Log-rank test {survfit2_p(km.AB)}"),
x = "Time in months",
y = "Cumulative Incidence") +
theme(panel.grid.major.y = element_line(linewidth = 0.02, color = "grey80"),
legend.position = c(0.85, 0.89))34.5.2 The log-Rank test
The log-rank test is a non-parametric method for comparing two or more survival curves. It examines the distribution of the entire survival time rather than comparing the survival probability at specific time points.
The log-rank test can be used to test the global null hypothesis that the curves are the same.
#Run log-Rank test to compare the curves for the intervention groups A Vs B
SurvDiff <- survdiff(Surv(time, status) ~ intervention, data = dat)
SurvDiffCall:
survdiff(formula = Surv(time, status) ~ intervention, data = dat)
N Observed Expected (O-E)^2/E (O-E)^2/V
intervention=therapy A (new) 21 9 17.65 4.24 13.3
intervention=therapy B (standard) 21 18 9.35 8.00 13.3
Chisq= 13.3 on 1 degrees of freedom, p= 3e-04 The log-rank test yields a chi-square statistical value, as it is built upon this framework. Therefore, the log-rank estimates the expected number of deaths among patients and compares it with the actual number of observed deaths. The resulting p-value is 0.0003, which means that we reject the null hypothesis and that the survival curves are significantly different.
34.5.3 The Hazard Ratio (HR)
The Hazard ratio (HR) is an estimate of the ratio of the hazard rate in the new therapy versus the standard therapy. The hazard ratio is often interpreted as a risk ratio or odds ratio, although they are not technically the same.
Hazard ratios (HRs) differ from relative risks (RRs) and odds ratio (OR) in that RRs and ORs are cumulative over an entire study, using a defined endpoint, while HRs represent instantaneous risk over the study time period.
After computing the number of observed events (deaths) in each group (
We also can compute the 95% confidence interval for the HR. First, we compute the standard error of the natural logarithm of the hazard ratio:
Second, we calculate the 95% CI of the natural logarithm of the hazard ratio. The lower limit is:
and the upper limit:
Finally, we convert each of these values back to HR using:
The hazard of dying is 74% lower (0.26 - 1 = -0.74) in the new treatment than in the standard treatment (HR=0.26; 95% CI: 0.12, 0.58).
In R:
The HR is:
O_A <- SurvDiff$obs[1]
O_B <- SurvDiff$obs[2]
E_A <- SurvDiff$exp[1]
E_B <- SurvDiff$exp[2]
HR <- (O_A/E_A)/(O_B/E_B)
HR[1] 0.2649746and the standard error of the logarithm of HR:
SE_lnHR = sqrt(1/E_A + 1/E_B)
SE_lnHR[1] 0.4044623The lower and upper limits of the logarithm of HR are:
LL_lnHR <- log(HR) - 1.96*SE_lnHR
LL_lnHR[1] -2.120867UL_lnHR <- log(HR) + 1.96*SE_lnHR
UL_lnHR[1] -0.5353751Finally: