24 One-way ANOVA test
One-way analysis of variance, usually referred to as one-way ANOVA, is a statistical test used when we want to compare several means. We may think of it as an extension of Student’s t-test to the case of more than two independent samples.
Although, this test can detect a difference between several groups it does not inform us about which groups are different from the others. At first glance, we might think to compare all groups in pairs with multiple t-tests. However, this procedure may lead to incorrect conclusions (known as multiple comparisons problem) because each comparison increases the likelihood of committing at least one Type I error within a set of comparisons (familly-wise Type I error rate).
This is the reason why, after an ANOVA test concluding on a significant difference between group means, we should not just compare all possible pairs of groups with t-tests. Instead we perform statistical tests that take into account the number of planned comparisons (post hoc tests) and make the necessary adjustments to ensure that Type I error is not inflated.
When we have finished this Chapter, we should be able to:
24.1 Research question and Hypothesis Testing
We consider the data in dataDWL dataset. In this example we explore the variations between weight loss according to four different types of diet. The question that may be asked is: does the average weight loss differ according to the diet?
24.2 Packages we need
We need to load the following packages:
24.3 Preparing the data
We import the data dataDWL in R:
library(readxl)
dataDWL <- read_excel(here("data", "dataDWL.xlsx"))We inspect the data and the type of variables:
glimpse(dataDWL)Rows: 60
Columns: 2
$ WeightLoss <dbl> 9.9, 9.6, 8.0, 4.9, 10.2, 9.0, 9.8, 10.8, 6.2, 8.3, 12.9, 1…
$ Diet <chr> "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A",…The dataset dataDWL has 60 participants and includes two variables. The numeric (<dbl>) WeightLoss variable and the character (<chr>) Diet variable (with levels “A”, “B”, “C” and “D”) which should be converted to a factor variable using the factor() function as follows:
24.4 Assumptions
A. Explore the characteristics of distribution for each group and check for normality
The distributions can be explored visually with appropriate plots. Additionally, summary statistics and significance tests to check for normality (e.g., Shapiro-Wilk test) can be used.
Graphs
We can visualize the distribution of WeightLoss for the four Diet groups:
set.seed(123)
ggplot(dataDWL, aes(x=Diet, y=WeightLoss)) +
geom_flat_violin(aes(fill = Diet), scale = "count") +
geom_boxplot(width = 0.14, outlier.shape = NA, alpha = 0.5) +
geom_point(position = position_jitter(width = 0.05),
size = 1.2, alpha = 0.6) +
ggsci::scale_fill_jco() +
theme_classic(base_size = 14) +
theme(legend.position="none",
axis.text = element_text(size = 14))The above figure shows that the data are close to symmetry and the assumption of a normal distribution is reasonable. Additionally, we can observe that the largest weight loss seems to have been achieved by the participants in C diet.
Summary statistics
The WeightLoss summary statistics for each diet group are:
DWL_summary <- dataDWL %>%
group_by(Diet) %>%
dplyr::summarise(
n = n(),
na = sum(is.na(WeightLoss)),
min = min(WeightLoss, na.rm = TRUE),
q1 = quantile(WeightLoss, 0.25, na.rm = TRUE),
median = quantile(WeightLoss, 0.5, na.rm = TRUE),
q3 = quantile(WeightLoss, 0.75, na.rm = TRUE),
max = max(WeightLoss, na.rm = TRUE),
mean = mean(WeightLoss, na.rm = TRUE),
sd = sd(WeightLoss, na.rm = TRUE),
skewness = EnvStats::skewness(WeightLoss, na.rm = TRUE),
kurtosis= EnvStats::kurtosis(WeightLoss, na.rm = TRUE)
) %>%
ungroup()
DWL_summary# A tibble: 4 × 12
Diet n na min q1 median q3 max mean sd skewness kurtosis
<fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A 15 0 4.9 8.15 9.6 10.5 12.9 9.18 2.30 -0.471 -0.302
2 B 15 0 3.8 7.85 9.2 10.8 12.7 8.91 2.78 -0.467 -0.515
3 C 15 0 8.7 10.8 12.2 13 15.1 12.1 1.79 -0.0451 -0.530
4 D 15 0 5.8 9.5 10.5 11.8 13.7 10.5 2.23 -0.475 0.229dataDWL %>%
group_by(Diet) %>%
dlookr::describe(WeightLoss) %>%
select(described_variables, Diet, n, mean, sd, p25, p50, p75, skewness, kurtosis) %>%
ungroup()Registered S3 methods overwritten by 'dlookr':
method from
plot.transform scales
print.transform scales# A tibble: 4 × 10
described_variables Diet n mean sd p25 p50 p75 skewness
<chr> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 WeightLoss A 15 9.18 2.30 8.15 9.6 10.5 -0.471
2 WeightLoss B 15 8.91 2.78 7.85 9.2 10.8 -0.467
3 WeightLoss C 15 12.1 1.79 10.8 12.2 13 -0.0451
4 WeightLoss D 15 10.5 2.23 9.5 10.5 11.8 -0.475
# ℹ 1 more variable: kurtosis <dbl>The means are close to medians and the standard deviations are also similar. Moreover, both skewness and (excess) kurtosis falls into the acceptable range of [-1, 1] indicating approximately normal distributions for all diet groups.
Normality test
The Shapiro-Wilk test for normality for each diet group is:
# A tibble: 4 × 4
Diet variable statistic p
<fct> <chr> <dbl> <dbl>
1 A WeightLoss 0.958 0.662
2 B WeightLoss 0.941 0.390
3 C WeightLoss 0.964 0.768
4 D WeightLoss 0.944 0.435The tests of normality suggest that the data for the WeightLoss in all groups are normally distributed (p > 0.05).
B. Levene’s test for equality of variances
The Levene’s test for equality of variances is:
dataDWL %>%
levene_test(WeightLoss ~ Diet)# A tibble: 1 × 4
df1 df2 statistic p
<int> <int> <dbl> <dbl>
1 3 56 0.600 0.617Since the p = 0.617 > 0.05, the null hypothesis (
24.5 Run the one-way ANOVA test
Now, we will perform an one-way ANOVA (with equal variances: Fisher’s classic ANOVA) to test the null hypothesis that the mean weight loss is the same for all the diet groups.
dataDWL %>%
anova_test(WeightLoss ~ Diet, detailed = T)ANOVA Table (type II tests)
Effect SSn SSd DFn DFd F p p<.05 ges
1 Diet 97.33 296.987 3 56 6.118 0.001 * 0.247The statistic F=6.118 indicates the obtained F-statistic = (variation between sample means
The p=0.001 is lower than 0.05. There is at least one diet with mean weight loss which is different from the others means.
From ANOVA table provided by the {rstatix} we can also calculate the generalized effect size (ges). The ges is the proportion of variability explained by the factor Diet (SSn) to total variability of the dependent variable (SSn + SSd), so:
ges of 0.247 (24.7%) means that 24.7% of the change in the weight loss can be accounted for the diet conditions.
Present the results in a summary table
A summary table can also be presented:
Show the code
| Characteristic | A, N = 151 | B, N = 151 | C, N = 151 | D, N = 151 | p-value2 |
|---|---|---|---|---|---|
| Weight Loss (kg) | 9.2 (2.3) | 8.9 (2.8) | 12.1 (1.8) | 10.5 (2.2) | 0.001 |
| 1 Mean (SD) | |||||
| 2 One-way ANOVA | |||||
24.6 Post-hoc tests
A significant one-way ANOVA is generally followed up by post-hoc tests to perform multiple pairwise comparisons between groups:
It is appropriate to use this test when one desires all the possible comparisons between a large set of means (e.g., 6 or more means) and the variances are supposed to be equal.
# A tibble: 6 × 9
term group1 group2 null.value estimate conf.low conf.high p.adj
* <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Diet A B 0 -0.273 -2.50 1.95 0.988
2 Diet A C 0 2.93 0.707 5.16 0.00513
3 Diet A D 0 1.36 -0.867 3.59 0.377
4 Diet B C 0 3.21 0.980 5.43 0.0019
5 Diet B D 0 1.63 -0.593 3.86 0.222
6 Diet C D 0 -1.57 -3.80 0.653 0.252
# ℹ 1 more variable: p.adj.signif <chr>The output contains the following columns of interest:
- estimate: estimate of the difference between means of the two groups
- conf.low, conf.high: the lower and the upper end point of the confidence interval at 95% (default)
- p.adj: p-value after adjustment for the multiple comparisons.
two_groups_unpaired <- load(dataDWL,
x = Diet, y = WeightLoss,
idx = c("C", "A", "B", "D")
)
two_groups_unpaired.mean_diff <- mean_diff(two_groups_unpaired)
dabest_plot(two_groups_unpaired.mean_diff)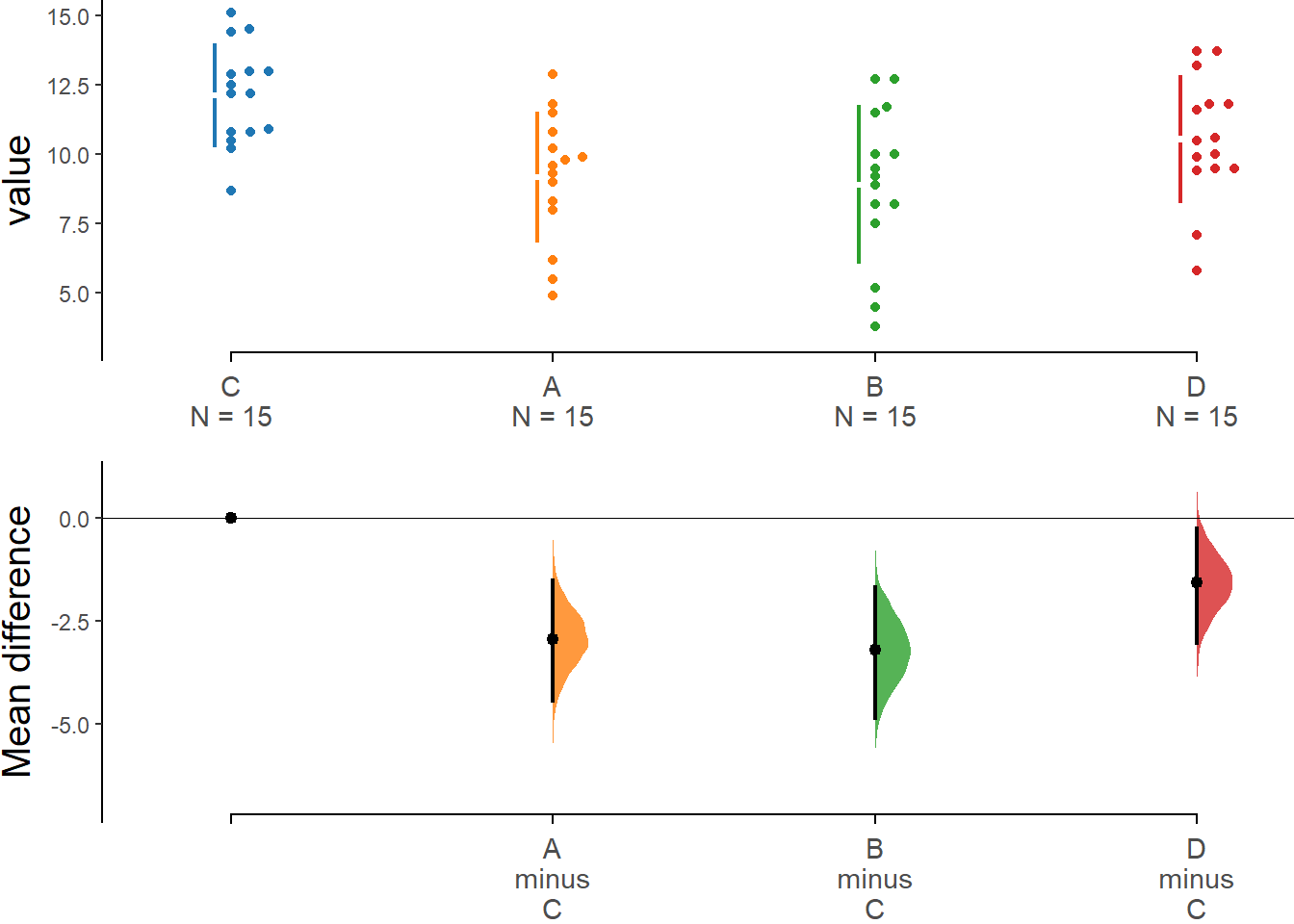
Alternatively, we can perform pairwise comparisons using pairwise t-test with the assumption of equal variances (pool.sd = TRUE) and calculate the adjusted p-values using Bonferroni correction:
pwc_Bonferroni <- dataDWL %>%
pairwise_t_test(
WeightLoss ~ Diet, pool.sd = TRUE,
p.adjust.method = "bonferroni"
)
pwc_Bonferroni # A tibble: 6 × 9
.y. group1 group2 n1 n2 p p.signif p.adj p.adj.signif
* <chr> <chr> <chr> <int> <int> <dbl> <chr> <dbl> <chr>
1 WeightLoss A B 15 15 0.746 ns 1 ns
2 WeightLoss A C 15 15 0.000954 *** 0.00572 **
3 WeightLoss B C 15 15 0.000344 *** 0.00206 **
4 WeightLoss A D 15 15 0.111 ns 0.669 ns
5 WeightLoss B D 15 15 0.0571 ns 0.343 ns
6 WeightLoss C D 15 15 0.0666 ns 0.399 ns Pairwise comparisons were carried out using the method of Tukey (or Bonferroni) and the adjusted p-values were calculated.
The results in Tukey post hoc table show that the weight loss from diet C seems to be significantly larger than diet A (mean difference = 2.91 kg, 95%CI [0.71, 5.16], p=0.005 <0.05) and diet B (mean difference = 3.21 kg, 95%CI [0.98, 5.43], p=0.002 <0.05).
24.7 Welch one-way ANOVA
If the variance is different between the groups (unequal variances) then the degrees of freedom associated with the ANOVA test are calculated differently (Welch one-way ANOVA).
# Welch one-way ANOVA test (not assuming equal variance)
dataDWL %>%
welch_anova_test(WeightLoss ~ Diet)# A tibble: 1 × 7
.y. n statistic DFn DFd p method
* <chr> <int> <dbl> <dbl> <dbl> <dbl> <chr>
1 WeightLoss 60 7.02 3 30.8 0.000989 Welch ANOVAIn this case, the Games-Howell post hoc test (or pairwise t-tests with no assumption of equal variances with Bonferroni correction) can be used to compare all possible combinations of group differences.
Games-Howell post hoc test
# Pairwise comparisons (Games-Howell)
pwc_GH <- dataDWL %>%
games_howell_test(WeightLoss ~ Diet)
pwc_GH# A tibble: 6 × 8
.y. group1 group2 estimate conf.low conf.high p.adj p.adj.signif
* <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
1 WeightLoss A B -0.273 -2.82 2.28 0.991 ns
2 WeightLoss A C 2.93 0.872 4.99 0.003 **
3 WeightLoss A D 1.36 -0.898 3.62 0.371 ns
4 WeightLoss B C 3.21 0.849 5.56 0.005 **
5 WeightLoss B D 1.63 -0.889 4.16 0.308 ns
6 WeightLoss C D -1.57 -3.60 0.452 0.17 ns 